Chapter 4 Analysis
4.1 Mutational Signature Analysis for User-uploaded data
Due to the ubiquitous nature of many of the signatures found across different cancer types, researchers may be interested in interrogating the presence and prevalence in their collected tumor samples. However, the analysis of cancer genomic data requires advanced bioinformatics skills and remains limited to a small research community. Accordingly, mSignatureDB provides user-freindly web interfaces for two popular mutational signature analysis tools, the 1.deconstructSigs and 2. the WTSI Mutational Signature Framework, to make such analyses more accessible to a wider community of researchers.
Results will be maintained for a month
4.1.1 Supported input file formats
1. Mutational Annotation Format (MAF): The format of a MAF file is tab-delimited columns (Column specification).
six minimum required columns:
- Chromosome
- Start_Position
- End_Position
- Reference_Allele
- Tumor_Seq_Allele2
- Tumor_Sample_Barcode
2. ICGC Simple Somatic Mutation file (TSV)
To make data publically availabe, the International Cancer Genome Consortium (ICGC) provides mutational profile of each Cancer project in Simple Somatic Mutation Format, which is similar to MAF in tab-delimited format. However, the field names and classification of variants are different from that of MAF. An example of ICGC TSV file can refer to this link.
Detailed instruction on ICGC Simple Somatic Mutation Format can refer to ICGC DCC Docs.
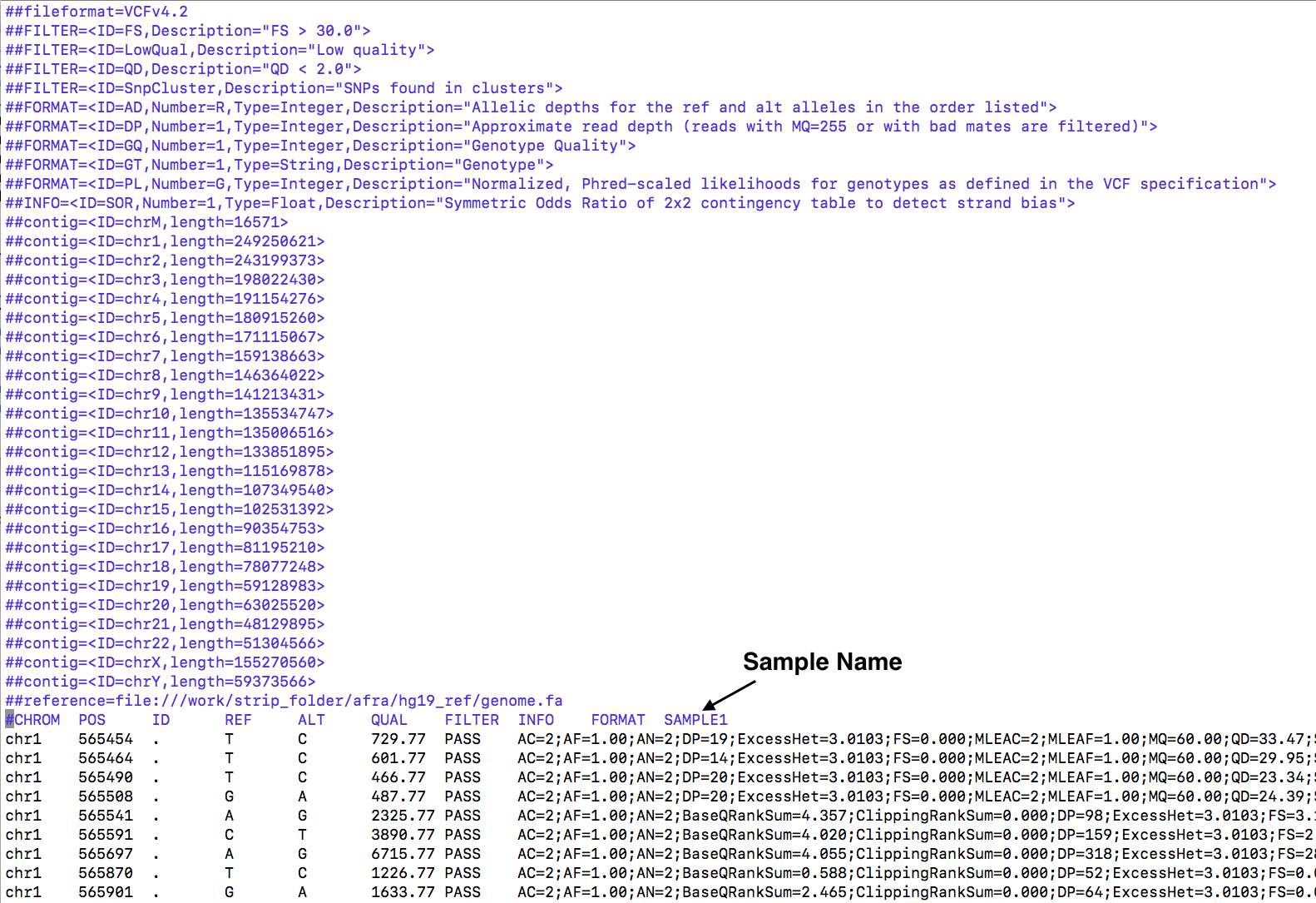
3. Variant Call Format (VCF)
Single sample VCF file is supported.
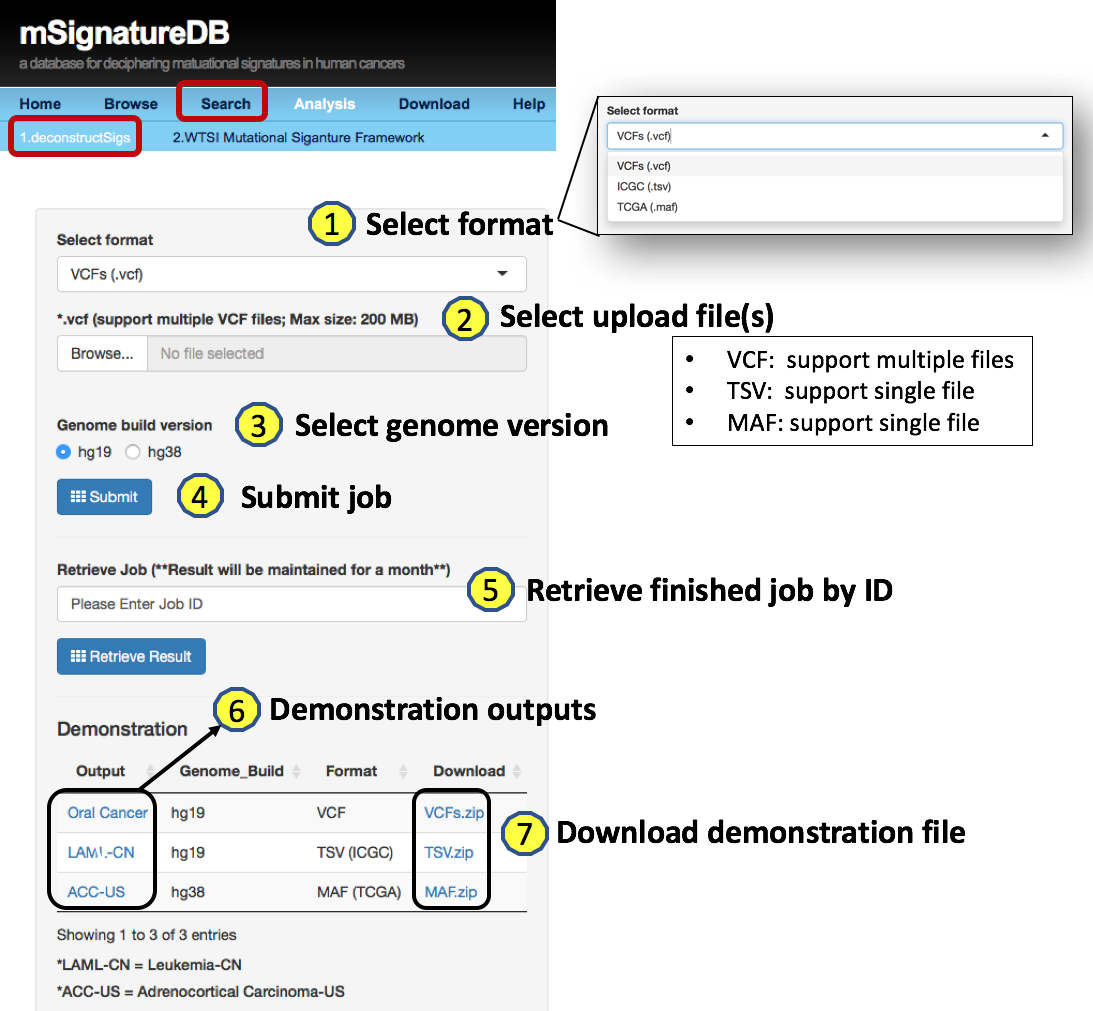
4.1.2 Supported genome build version
- hg19
- hg38
4.2 Web-based deconstructSigs
The deconstructSigs method is suitable for analyzing the contribution of known COSMIC signatures in study cohorts with small number of samples, which has been proved to be able to consistently identify the same signatures of mutational processes active in a single tumor sample compared with the analysis of an entire sample set using the WTSI Mutational Signature Framework.
4.2.1 Input
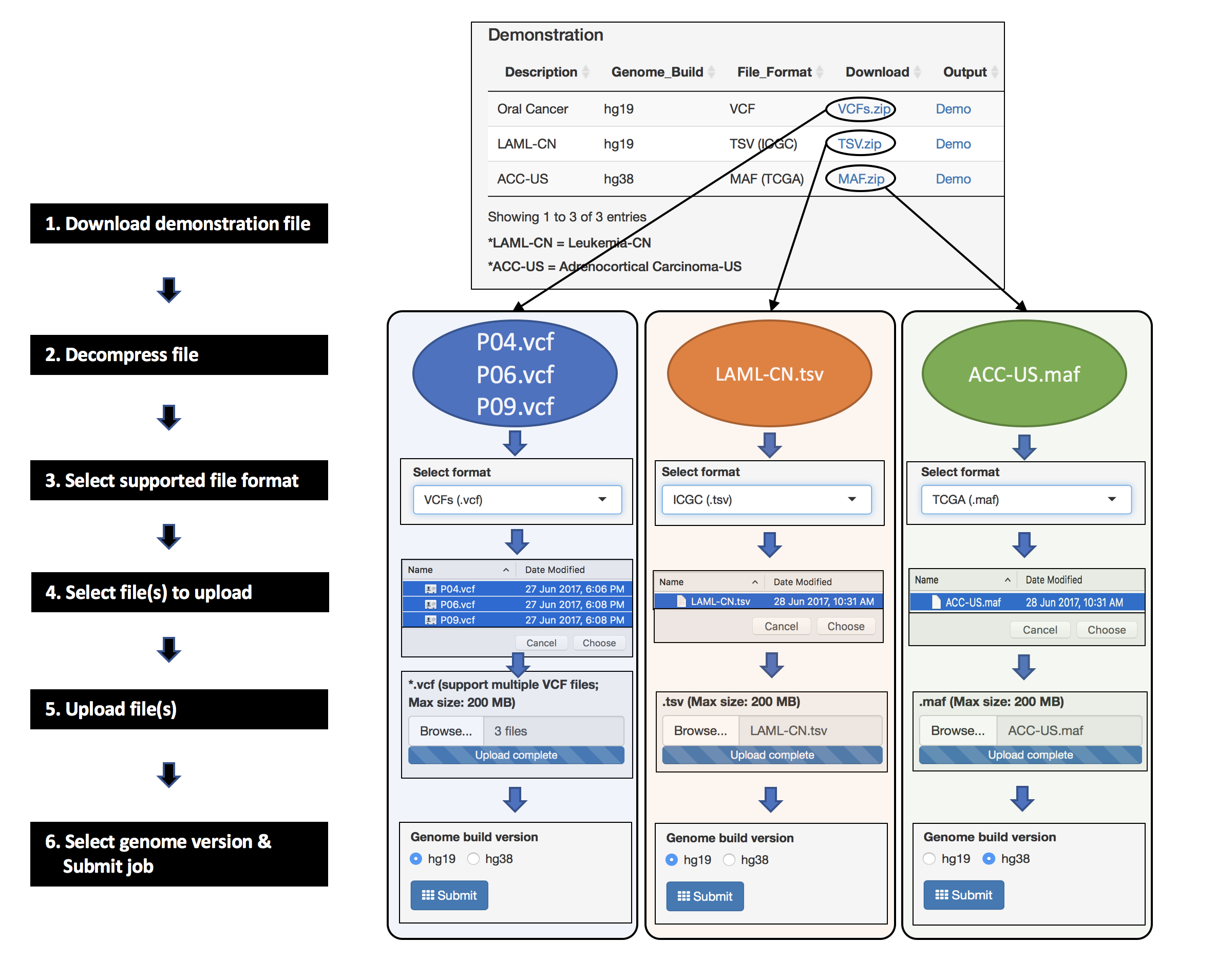
4.2.2 Output
4.2.2.1 Mutational Signature analysis result at Project level
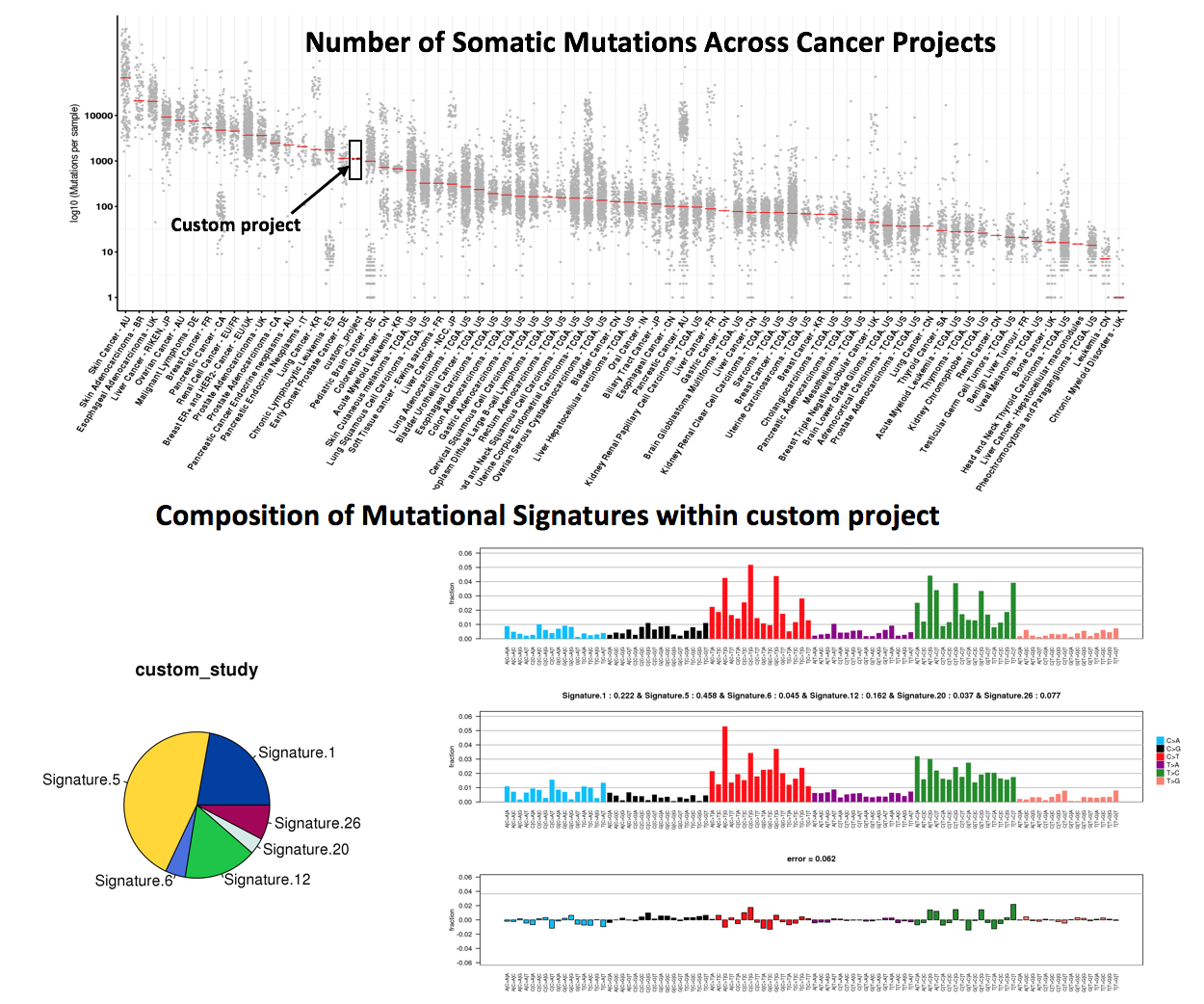
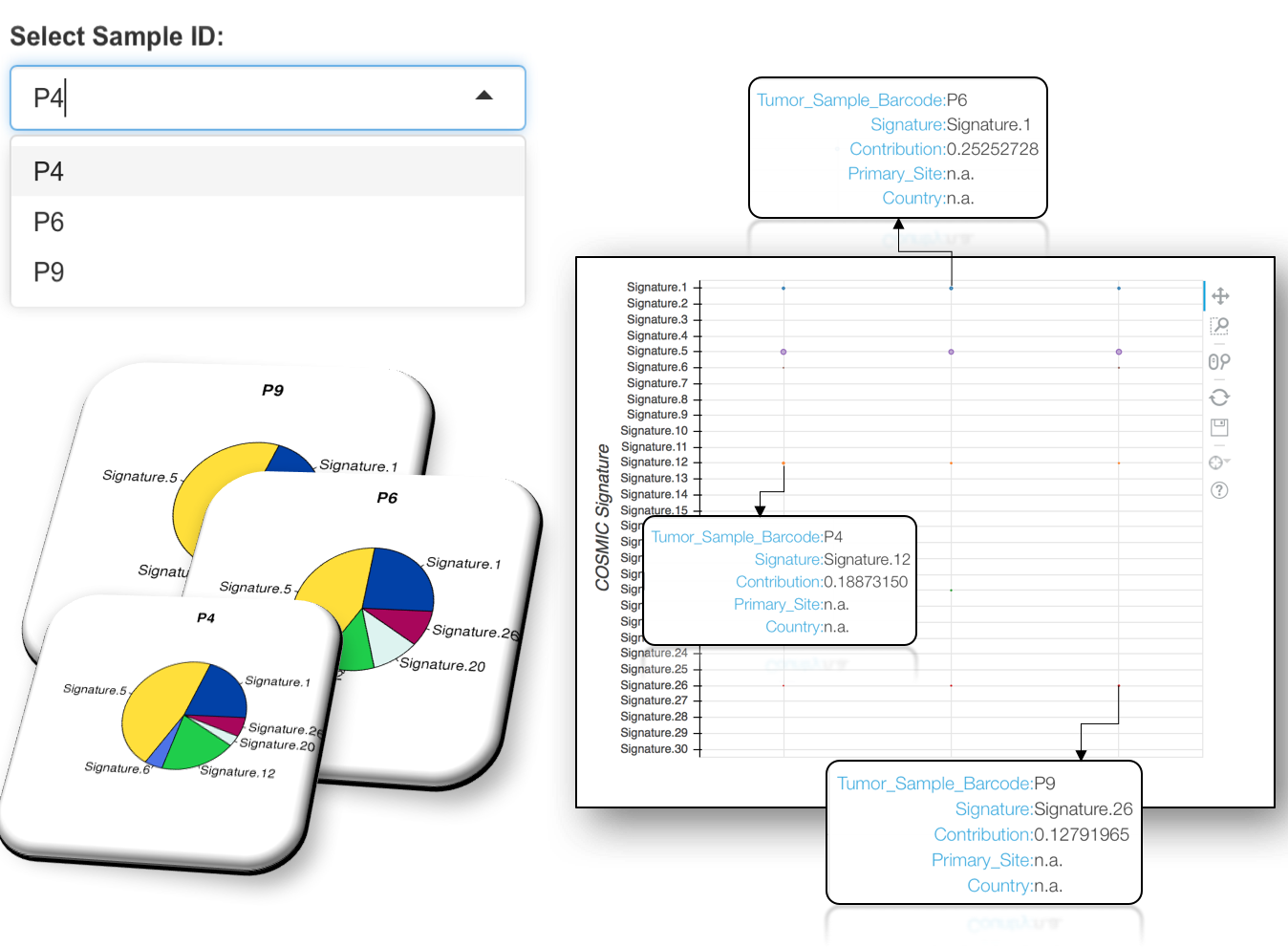
4.2.2.2 Mutational Signature analysis result at Sample resolution
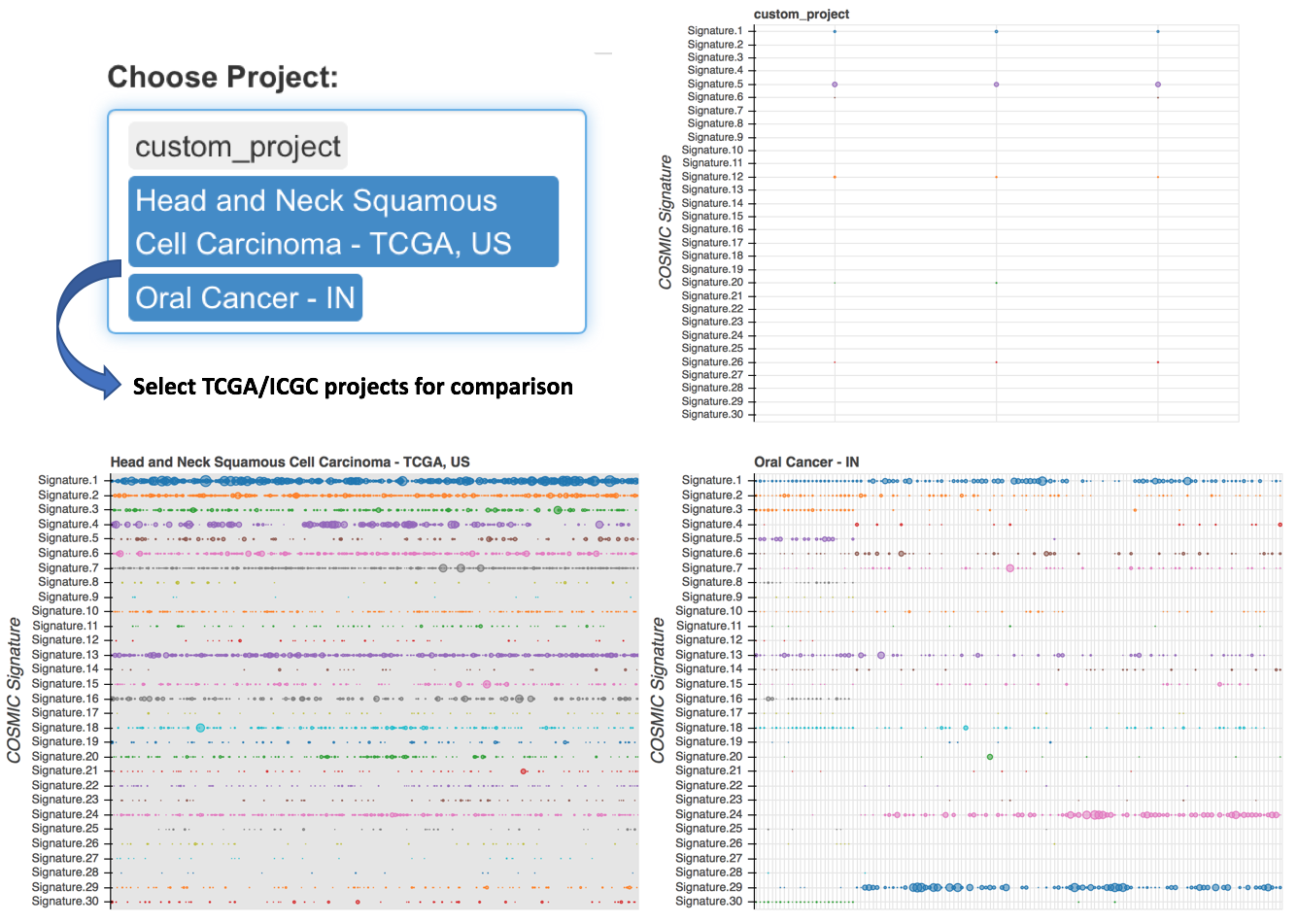
4.2.2.3 Cross project comparsion
4.3 Web-based WTSI Mutational Signature Framework
The Wellcome Trust Sanger Institute (WTSI) Mutational Signature Framework is recommended for identifying novel mutational signatures when large samples are available. As suggested by previous study, at least 200 cancer genome catalogs are required for accurately decomposing signatures of 20 mutational processes. The original WTSI Mutational Signature Framework is developed on MATLAB, which requires a commercial license and basic knowledge on MATLAB to perform the same analysis as Alexandrov et al. Accordingly, we incorperated the R-based implementation of the MATLAB WTSI framework and provided a web interface for this framework, which may be beneficial for mSignatureDB users.
4.3.1 Input
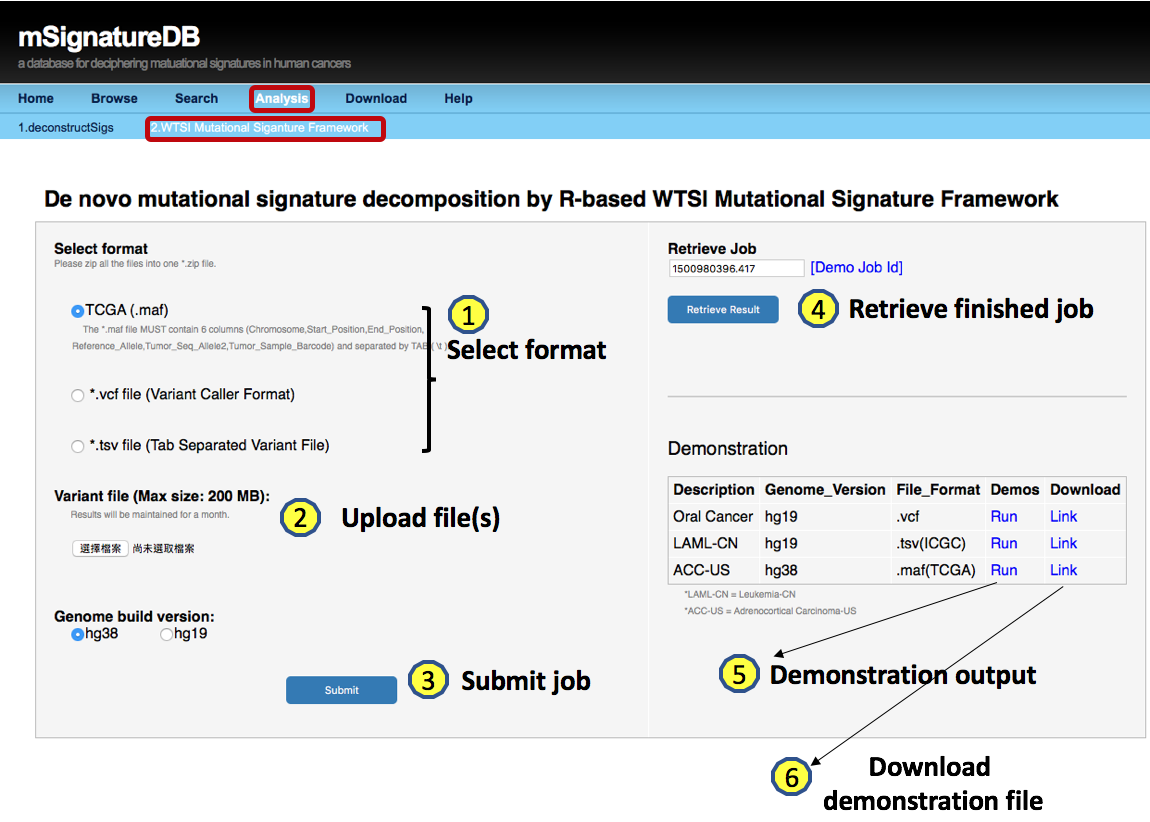
4.3.2 Output
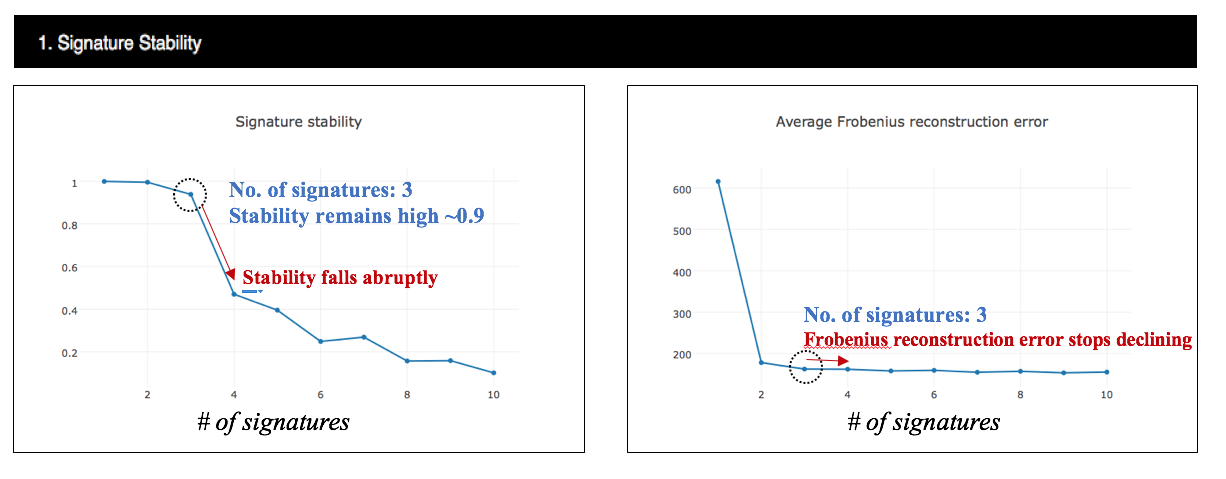
4.3.2.1 Estimate the number of mutational signatures present in the analysed dataset
Signature stability and the average Frobenius reconstruction error for the analysis of custom cohort. As shown in the following figure, signature stability remains high for 3 signatures extracted, then falls abruptly at the same point that frobenius reconstruction error stops declining. These plots indicate that three appears to be the optimal number and 3 mutational signatures can be stably identified.
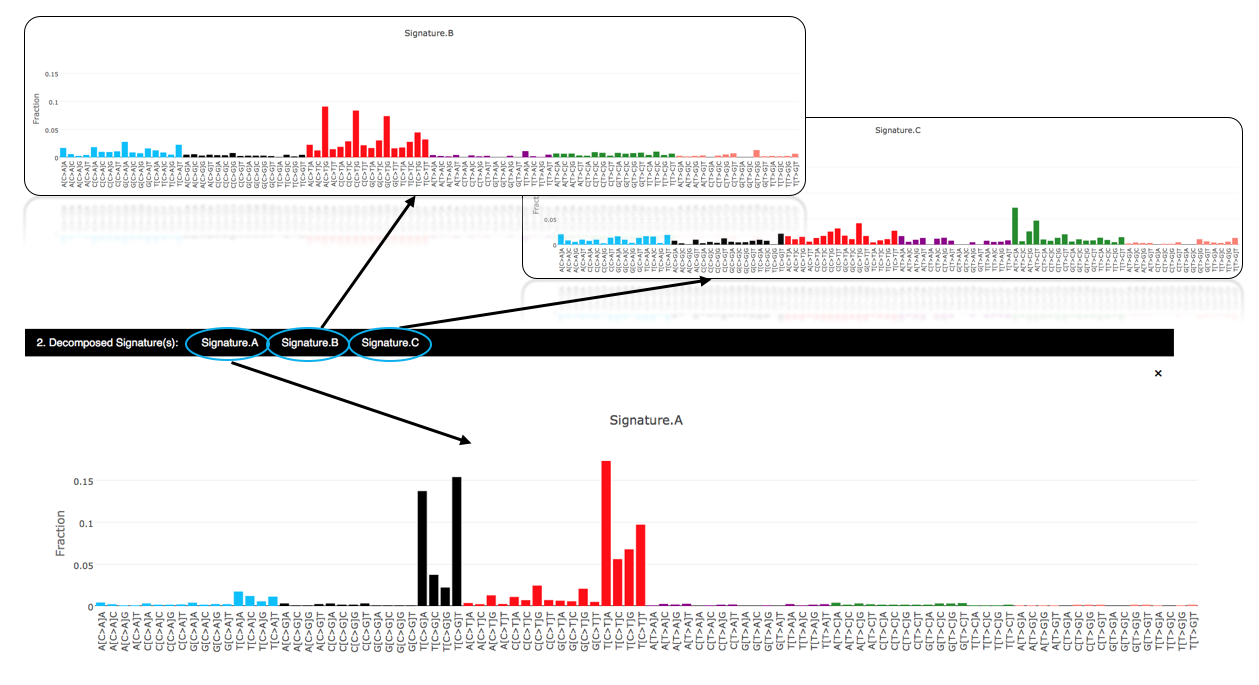
4.3.2.2 Decomposed signatures
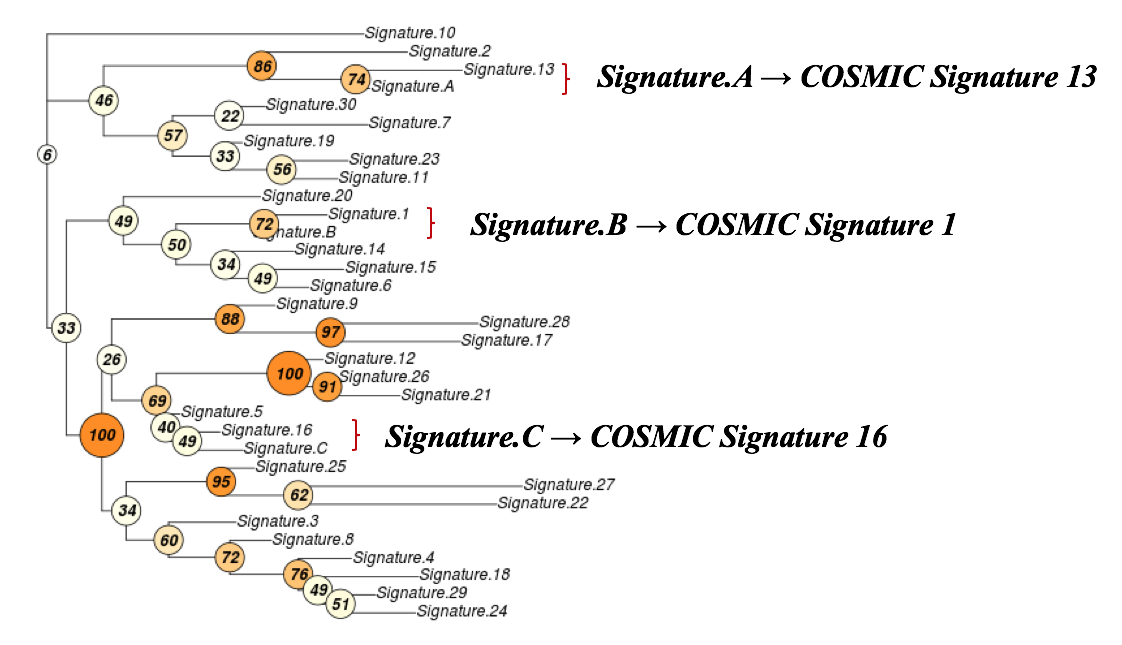
Judging from the estimation result of signature stability, a total of 3 signatures seems to explain the charateristics of the mutational spectrum well. The user-uploaded mutational profile can be decomposed into 3 signatures: Signature.A, Signature.B, and Signature.C. The fraction of mutations in each of 96 trinucleotide contexts can be further inspected through the hyperlinks embbeded in the output page.
4.3.2.3 The contibution of the 3 identified signatures in custom cohort
The pie chart shows the weights of each signature contributing to the custom cohort. 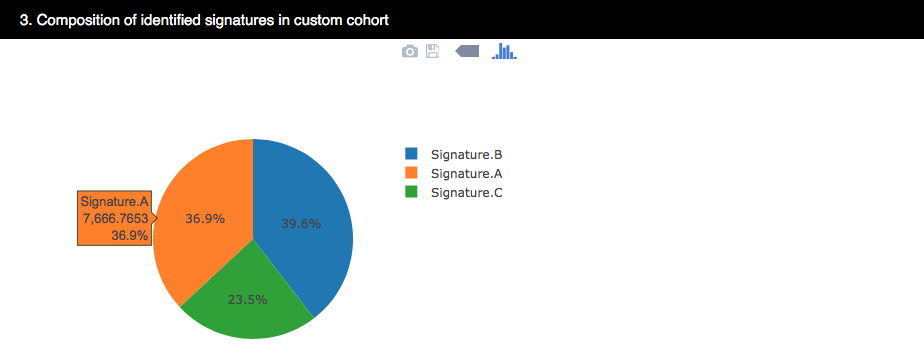
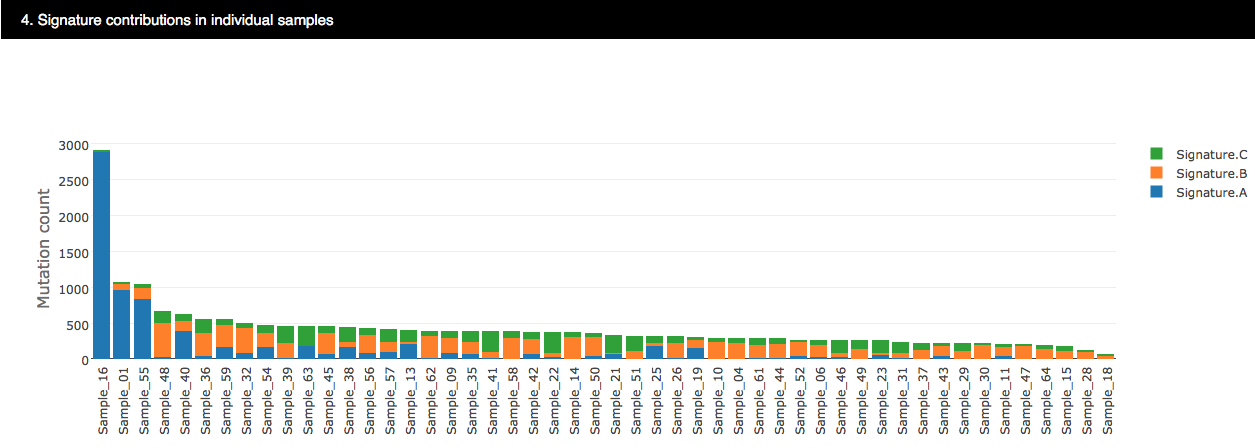
4.3.2.4 Signature compositions in individual samples
The bar chart shows the weights of each identified signature present in an individual sample.
4.3.2.5 Known signature assignment
Most of the existing signature analysis packages can perform NMF decomposition and extract mutational signatures from samples of a cohort. Signature assignment is the last step of de novo signature analysis, which can be easily achieved by cosine similarity analysis but is always neglected by existing signature analysis tools, making known signature assignment and novel signature identification very inconvenient. Accordingly, we incorperated the bootrapped cosine similarity function of the R supraHex package into mSignatureDB to facilitate known signature assignment and to calculate statistical significance of similarity between mutational signatures. As shown in the figure below, the decomposed signatures can be assigned to COSMIC Signaure 13, 1 and 16, respectively.