mSignatureDB Help
Po-Jung Huang
Oct. 1, 2017
Chapter 1 Introduction
1.1 Overview of mSignatureDB
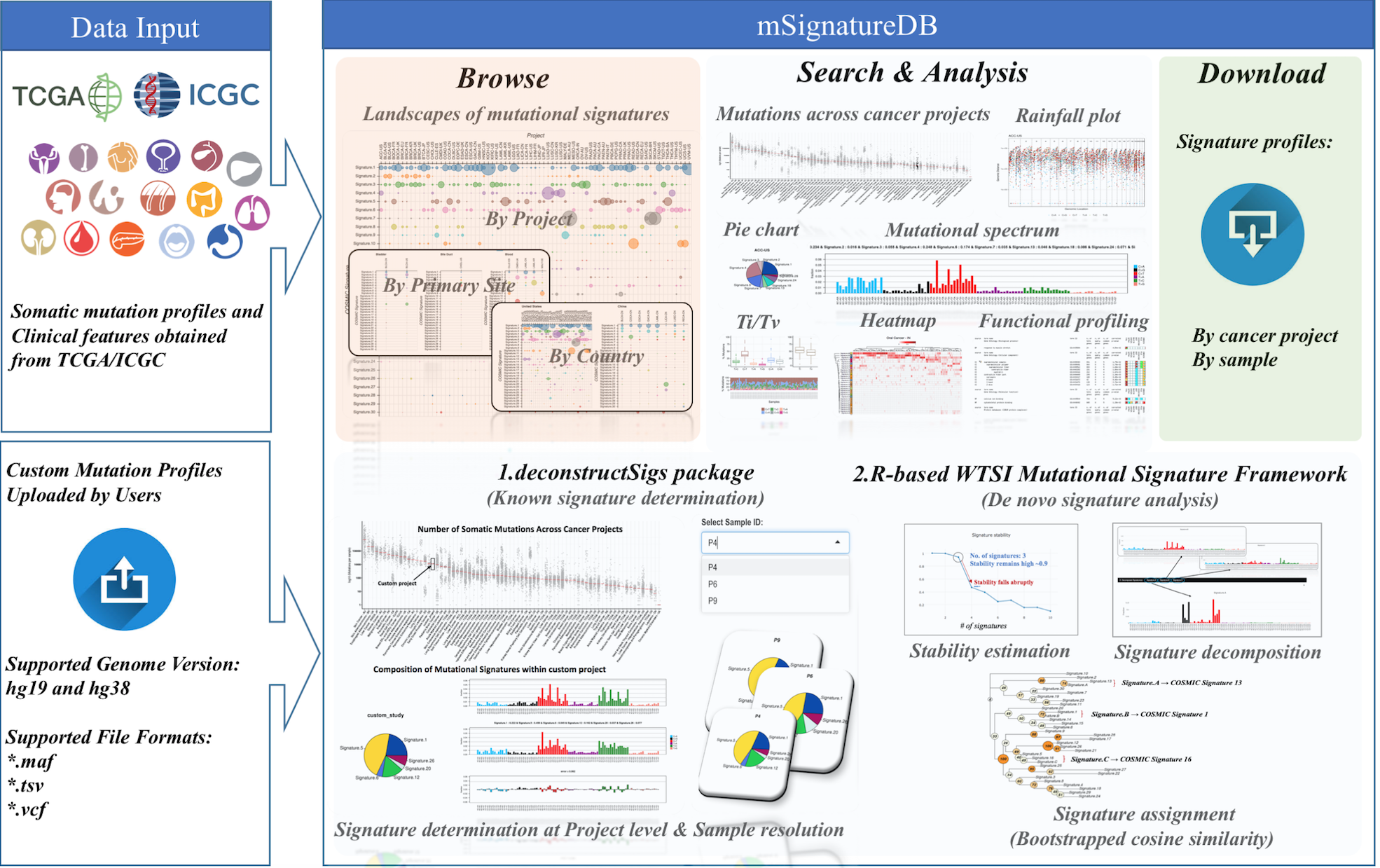
1.2 Main features of mSignatureDB
mSignatureDB comprises 4 major components: (1) Browse; (2) Search; (3) Analysis; and (4) Download.
(1) In the “Browse” page, the landscapes and contributions of mutational sigantures can be inspected and arranged according to project, primary site, country and individual sample.
(2) In the “Search” page, users can search the database using names of the TCGA/ICGC cancer projects. The contribution of each signature can be compared within and between cancer projects. The heatmap, word cloud generator, and bootstrapped clustering modules are used to reveal dominant signature patterns and to identify statistically significant differences between cancer projects and subsets of patients (e.g., gender, tumor stage, and vital status), respectively.
(3) In the “Analysis” page, mSignatureDB provides user-friendly interfaces for two popular mutational signature anlysis tools:
- deconstructSigs (for known sigantures identification)
- R-based WTSI Mutational Signature Framework (for de novo signature analysis),
to make such analyses more accessible to a wider community of researchers.
(4) In the “Download” page, the mutational signature patterns (96 trinucleotide count matrices) of TCGA/ICGC tumors can be download for further inspection by project.